Decision trees in python again, cross-validation
Jun 25, 2015
warning
This post is more than 5 years old. While math doesn't age, code and operating systems do. Please use the code/ideas with caution and expect some issues due to the age of the content. I am keeping these posts up for archival purposes because I still find them useful for reference, even when they are out of date!
This is my second post on decision trees using scikit-learn and Python. The first, Decision trees in python with scikit-learn and pandas focused on visualizing the resulting tree. This post will concentrate on using cross-validation methods to choose the parameters used to train the tree. In particular, I'll focus on grid* and random search for decision tree parameter settings. If this sounds interesting to you, following along. As always, comments, questions and corrections are welcome below.
gist
Okay, let's get started by linking to the gist containing all of the needed code-- linked here. So, you don't have to copy and paste (unless you want to). The following code is adapted from this scikit-learn example (be careful-- although the example looks like it uses the iris data it really loads the digits data). Also, that example uses a random forest-- an ensemble of decision trees-- so the parameters that can be searched are slightly different.
imports
First we import all of the code we'll need for the post-- this expands on the methods needed in the last post:
from __future__ import print_function
import os
import subprocess
from time import time
from operator import itemgetter
from scipy.stats import randint
import pandas as pd
import numpy as np
from sklearn.tree import DecisionTreeClassifier
from sklearn.tree import export_graphviz
from sklearn.grid_search import GridSearchCV
from sklearn.grid_search import RandomizedSearchCV
from sklearn.cross_validation import cross_val_score
The main additions are methods from
sklearn.grid_search as well as some
tools to:
-
time the searches, using
time, -
sort the results, using
itemgetter, and -
generate random integers, using
scipy.stats.randint.
Now we can start writing our functions-- some new, some old.
previous functions
I will also declare functions used in the previous post so that I can use them here. These include:
-
get_code-- writes pseudo-code for a decision tree, -
visualize_tree-- to generate a graphic of a decision tree. The ability to name output files has been added here. -
encode_target-- process raw data for use with scikit-learn. -
get_iris_data-- grabs iris.csv from the web, if needed, and writes a copy to the local directory. This is mainly to replicate real-world usage of pandas and scikit-learn.
def get_code(tree, feature_names, target_names,
spacer_base=" "):
"""Produce pseudo-code for decision tree.
Args
----
tree -- scikit-leant Decision Tree.
feature_names -- list of feature names.
target_names -- list of target (class) names.
spacer_base -- used for spacing code (default: " ").
Notes
-----
based on http://stackoverflow.com/a/30104792.
"""
left = tree.tree_.children_left
right = tree.tree_.children_right
threshold = tree.tree_.threshold
features = [feature_names[i] for i in tree.tree_.feature]
value = tree.tree_.value
def recurse(left, right, threshold, features, node, depth):
spacer = spacer_base * depth
if (threshold[node] != -2):
print(spacer + "if ( " + features[node] + " <= " + \
str(threshold[node]) + " ) {")
if left[node] != -1:
recurse (left, right, threshold, features,
left[node], depth+1)
print(spacer + "}\n" + spacer +"else {")
if right[node] != -1:
recurse (left, right, threshold, features,
right[node], depth+1)
print(spacer + "}")
else:
target = value[node]
for i, v in zip(np.nonzero(target)[1],
target[np.nonzero(target)]):
target_name = target_names[i]
target_count = int(v)
print(spacer + "return " + str(target_name) + \
" ( " + str(target_count) + " examples )")
recurse(left, right, threshold, features, 0, 0)
def visualize_tree(tree, feature_names, fn="dt"):
"""Create tree png using graphviz.
Args
----
tree -- scikit-learn Decision Tree.
feature_names -- list of feature names.
fn -- [string], root of filename, default `dt`.
"""
dotfile = fn + ".dot"
pngfile = fn + ".png"
with open(dotfile, 'w') as f:
export_graphviz(tree, out_file=f,
feature_names=feature_names)
command = ["dot", "-Tpng", dotfile, "-o", pngfile]
try:
subprocess.check_call(command)
except:
exit("Could not run dot, ie graphviz, "
"to produce visualization")
def encode_target(df, target_column):
"""Add column to df with integers for the target.
Args
----
df -- pandas Data Frame.
target_column -- column to map to int, producing new
Target column.
Returns
-------
df -- modified Data Frame.
targets -- list of target names.
"""
df_mod = df.copy()
targets = df_mod[target_column].unique()
map_to_int = {name: n for n, name in enumerate(targets)}
df_mod["Target"] = df_mod[target_column].replace(map_to_int)
return (df_mod, targets)
def get_iris_data():
"""Get the iris data, from local csv or pandas repo."""
if os.path.exists("iris.csv"):
print("-- iris.csv found locally")
df = pd.read_csv("iris.csv", index_col=0)
else:
print("-- trying to download from github")
fn = ("https://raw.githubusercontent.com/pydata/"
"pandas/master/pandas/tests/data/iris.csv")
try:
df = pd.read_csv(fn)
except:
exit("-- Unable to download iris.csv")
with open("iris.csv", 'w') as f:
print("-- writing to local iris.csv file")
df.to_csv(f)
return dfnew functions
report
Next we add some new function to do the grid and random searches as well as
report on the top parameters found. First up is
report. This function
takes the output from the grid or random search, prints a report of the top
models and returns the best parameter setting.
def report(grid_scores, n_top=3):
"""Report top n_top parameters settings, default n_top=3.
Args
----
grid_scores -- output from grid or random search
n_top -- how many to report, of top models
Returns
-------
top_params -- [dict] top parameter settings found in
search
"""
top_scores = sorted(grid_scores,
key=itemgetter(1),
reverse=True)[:n_top]
for i, score in enumerate(top_scores):
print("Model with rank: {0}".format(i + 1))
print(("Mean validation score: "
"{0:.3f} (std: {1:.3f})").format(
score.mean_validation_score,
np.std(score.cv_validation_scores)))
print("Parameters: {0}".format(score.parameters))
print("")
return top_scores[0].parametersgrid search
Next up is run_gridsearch. This
function takes
-
the features
X, -
the targets
y, -
a (Decision Tree) classifier
clf, -
a dictionary of parameters to try
param_grid -
the fold of the cross-validation
cv, defaulted to 5-- this is discussed more below.
The param_grid is the set of parameters
that will be tested-- be careful not to list too many options, because all
combinations will be tested!
def run_gridsearch(X, y, clf, param_grid, cv=5):
"""Run a grid search for best Decision Tree parameters.
Args
----
X -- features
y -- targets (classes)
cf -- scikit-learn Decision Tree
param_grid -- [dict] parameter settings to test
cv -- fold of cross-validation, default 5
Returns
-------
top_params -- [dict] from report()
"""
grid_search = GridSearchCV(clf,
param_grid=param_grid,
cv=cv)
start = time()
grid_search.fit(X, y)
print(("\nGridSearchCV took {:.2f} "
"seconds for {:d} candidate "
"parameter settings.").format(time() - start,
len(grid_search.grid_scores_)))
top_params = report(grid_search.grid_scores_, 3)
return top_paramsrandom search
Next up is the function run_randomsearch,
which samples parameters from
specified lists or distributions. Similar to the grid search, the arguments
are:
-
the features
X -
the targets
y -
a (Decision Tree) classifier
clf -
the fold of the cross-validation
cv, defaulted to 5-- this is discussed more below -
the number of random parameter setting to consider
n_iter_search, defaulted to 20.
def run_randomsearch(X, y, clf, para_dist, cv=5,
n_iter_search=20):
"""Run a random search for best Decision Tree parameters.
Args
----
X -- features
y -- targets (classes)
cf -- scikit-learn Decision Tree
param_dist -- [dict] list, distributions of parameters
to sample
cv -- fold of cross-validation, default 5
n_iter_search -- number of random parameter sets to try,
default 20.
Returns
-------
top_params -- [dict] from report()
"""
random_search = RandomizedSearchCV(clf,
param_distributions=param_dist,
n_iter=n_iter_search)
start = time()
random_search.fit(X, y)
print(("\nRandomizedSearchCV took {:.2f} seconds "
"for {:d} candidates parameter "
"settings.").format((time() - start),
n_iter_search))
top_params = report(random_search.grid_scores_, 3)
return top_paramsOkay, we've defined all our functions-- let's use them!
cross-validation
getting the data
Next, let's run through finding good parameter settings, using the search methods that we've setup above. First some preliminaries-- get the data and construct the feature and target data:
print("\n-- get data:")
df = get_iris_data()
print("")
features = ["SepalLength", "SepalWidth",
"PetalLength", "PetalWidth"]
df, targets = encode_target(df, "Name")
y = df["Target"]
X = df[features]producing...
-- get data:
-- iris.csv found locallya first cross-validation
Next, let's do cross-validation using the parameters from the previous post-- Decision trees in python with scikit-learn and pandas . I'll use 10-fold cross-validation in all of the examples to follow. This choice means:
- split the data into 10 parts
- fit on 9-parts
- test accuracy on the remaining part
This is repeated on all combinations to produce ten estimates of the accuracy of the model using the current parameter setting. Typically the mean and standard deviation of the ten scores is reported. So, if we use the setting from the previous post, we get:
print("-- 10-fold cross-validation "
"[using setup from previous post]")
dt_old = DecisionTreeClassifier(min_samples_split=20,
random_state=99)
dt_old.fit(X, y)
scores = cross_val_score(dt_old, X, y, cv=10)
print("mean: {:.3f} (std: {:.3f})".format(scores.mean(),
scores.std()),
end="\n\n" )producing...
-- 10-fold cross-validation [using setup from previous post]
mean: 0.960 (std: 0.033)Okay, 0.960 is not bad. That means that the average accuracy (percentage of correct classifications using the trained model) is 96%. That accuracy is pretty high, but let's if see if better parameters can be found.
application of grid search
First, I'll try a grid search. The dictionary
param_grid provides the
different parameter settings to test. The keys are the parameter name and the
values are a list of settings to try. Make sure to check the documentation
for Decision Trees (or other method you are testing) to get correct parameter
names and valid values to be tested.
print("-- Grid Parameter Search via 10-fold CV")
# set of parameters to test
param_grid = {"criterion": ["gini", "entropy"],
"min_samples_split": [2, 10, 20],
"max_depth": [None, 2, 5, 10],
"min_samples_leaf": [1, 5, 10],
"max_leaf_nodes": [None, 5, 10, 20],
}
dt = DecisionTreeClassifier()
ts_gs = run_gridsearch(X, y, dt, param_grid, cv=10)producing...
-- Grid Parameter Search via 10-fold CV
GridSearchCV took 5.02 seconds for 288 candidate parameter settings.
Model with rank: 1
Mean validation score: 0.967 (std: 0.033)
Parameters: {'min_samples_split': 10, 'max_leaf_nodes': 5,
'criterion': 'gini', 'max_depth': None, 'min_samples_leaf': 1}
Model with rank: 2
Mean validation score: 0.967 (std: 0.033)
Parameters: {'min_samples_split': 20, 'max_leaf_nodes': 5,
'criterion': 'gini', 'max_depth': None, 'min_samples_leaf': 1}
Model with rank: 3
Mean validation score: 0.967 (std: 0.033)
Parameters: {'min_samples_split': 10, 'max_leaf_nodes': 5,
'criterion': 'gini', 'max_depth': 5, 'min_samples_leaf': 1}
In most runs I get a mean of 0.967 for a variety of parameter settings. This
means there is an improvement from 96% to 96.7%-- I guess every bit helps! We
can see the best parameter setting ts_gs
as shown below:
print("\n-- Best Parameters:")
for k, v in ts_gs.items():
print("parameter: {:<20s} setting: {}".format(k, v))producing...
-- Best Parameters:
parameter: min_samples_split setting: 10
parameter: max_leaf_nodes setting: 5
parameter: criterion setting: gini
parameter: max_depth setting: None
parameter: min_samples_leaf setting: 1and, replicate the cross-validation results:
# test the retuned best parameters
print("\n\n-- Testing best parameters [Grid]...")
dt_ts_gs = DecisionTreeClassifier(**ts_gs)
scores = cross_val_score(dt_ts_gs, X, y, cv=10)
print("mean: {:.3f} (std: {:.3f})".format(scores.mean(),
scores.std()),
end="\n\n" )producing...
-- Testing best parameters [Grid]...
mean: 0.967 (std: 0.033)Next, let's use the code from the previous post (also provided above) to get psuedo-code for best tree:
print("\n-- get_code for best parameters [Grid]:", end="\n\n")
dt_ts_gs.fit(X,y)
get_code(dt_ts_gs, features, targets)producing...
-- get_code for best parameters [Grid]:
if ( PetalWidth <= 0.800000011921 ) {
return Iris-setosa ( 50 examples )
}
else {
if ( PetalWidth <= 1.75 ) {
if ( PetalLength <= 4.94999980927 ) {
if ( PetalWidth <= 1.65000009537 ) {
return Iris-versicolor ( 47 examples )
}
else {
return Iris-virginica ( 1 examples )
}
}
else {
return Iris-versicolor ( 2 examples )
return Iris-virginica ( 4 examples )
}
}
else {
return Iris-versicolor ( 1 examples )
return Iris-virginica ( 45 examples )
}
}We can also make a graphic of the Decision Tree:
visualize_tree(dt_ts_gs, features, fn="grid_best")resulting in
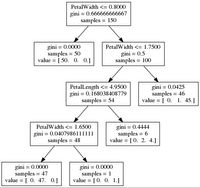
application of random search
Next, we try the random search method for finding parameters. In this case the
dictionary param_dist has keys
that are the parameter names (as
before) and values that are (i) a list to be sampled from, or (ii) a
distribution to be sampled from-- again, make sure the parameter names are
valid and the distributions produce values that are sensible for the method
being tested. In this example I use 288 samples so that the number of parameter
settings tested is the same as the grid search above:
print("-- Random Parameter Search via 10-fold CV")
# dict of parameter list/distributions to sample
param_dist = {"criterion": ["gini", "entropy"],
"min_samples_split": randint(1, 20),
"max_depth": randint(1, 20),
"min_samples_leaf": randint(1, 20),
"max_leaf_nodes": randint(2, 20)}
dt = DecisionTreeClassifier()
ts_rs = run_randomsearch(X, y, dt, param_dist, cv=10,
n_iter_search=288)producing...
-- Random Parameter Search via 10-fold CV
RandomizedSearchCV took 1.52 seconds for 288 candidates parameter
settings.
Model with rank: 1
Mean validation score: 0.967 (std: 0.033)
Parameters: {'min_samples_split': 12, 'max_leaf_nodes': 5,
'criterion': 'gini', 'max_depth': 19, 'min_samples_leaf': 1}
Model with rank: 2
Mean validation score: 0.967 (std: 0.033)
Parameters: {'min_samples_split': 1, 'max_leaf_nodes': 6, 'criterion':
'gini', 'max_depth': 11, 'min_samples_leaf': 1}
Model with rank: 3
Mean validation score: 0.967 (std: 0.033)
Parameters: {'min_samples_split': 4, 'max_leaf_nodes': 5, 'criterion':
'gini', 'max_depth': 15, 'min_samples_leaf': 1}As with the grid search, this typically finds multiple parameter settings with a mean accuracy 0.967, or 96.7%. As we did above, the parameters for the best cross-validation are:
print("\n-- Best Parameters:")
for k, v in ts_rs.items():
print("parameters: {:<20s} setting: {}".format(k, v))producing...
-- Best Parameters:
parameters: min_samples_split setting: 12
parameters: max_leaf_nodes setting: 5
parameters: criterion setting: gini
parameters: max_depth setting: 19
parameters: min_samples_leaf setting: 1And, we can test the best parameters again:
# test the retuned best parameters
print("\n\n-- Testing best parameters [Random]...")
dt_ts_rs = DecisionTreeClassifier(**ts_rs)
scores = cross_val_score(dt_ts_rs, X, y, cv=10)
print("mean: {:.3f} (std: {:.3f})".format(scores.mean(),
scores.std()),
end="\n\n" )producing...
-- Testing best parameters [Random]...
mean: 0.967 (std: 0.033)To see what the Decision Tree is like, we can generate the pseudo-code for best random search result
print("\n-- get_code for best parameters [Random]:")
dt_ts_rs.fit(X,y)
get_code(dt_ts_rs, features, targets)producing...
-- get_code for best parameters [Random]:
if ( PetalLength <= 2.45000004768 ) {
return Iris-setosa ( 50 examples )
}
else {
if ( PetalWidth <= 1.75 ) {
if ( PetalLength <= 4.94999980927 ) {
if ( PetalWidth <= 1.65000009537 ) {
return Iris-versicolor ( 47 examples )
}
else {
return Iris-virginica ( 1 examples )
}
}
else {
return Iris-versicolor ( 2 examples )
return Iris-virginica ( 4 examples )
}
}
else {
return Iris-versicolor ( 1 examples )
return Iris-virginica ( 45 examples )
}
}and visualize the tree
visualize_tree(dt_ts_rs, features, fn="rand_best")producing:
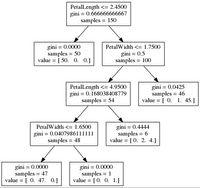
Wrapping Up
So, we've used grid and random search with cross-validation to tune the parameters for our Decision Tree. In both cases there were marginal improvements, from 96% to 96.7%. Of course, this effect can be much larger in more complicated problems. A few final notes:
- After finding the best parameter settings through cross-validation search it is typical to train the model with all of the data, using the best parameters found.
- The conventional wisdom is that random search is more efficient than grid search for practical application. This certainly makes sense in cases where there are many parameters and features to test and grid search is really (computationally) difficult-- takes too long.
- The basic cross-validation ideas developed here can be applied to many other scikit-learn models-- Random Forests, logistic regression, SVM, etc. To do this you simply need to spend some time learning about the model parameters, as well as the range of sensible values, to setup your own grid/random search-- I'd love to see it if you do this!
So, that's it. If you have comments or questions please leave them below-- links to code, web pages, etc are also very much appreciated by me and other readers. As always, reports of corrections/typos are also welcome. Enjoy!